How to Read Csv With No Header
Near of the data is available in a tabular format of CSV files. It is very popular. You can convert them to a pandas DataFrame using the read_csv function. The pandas.read_csv is used to load a CSV file equally a pandas dataframe.
In this article, you will learn the different features of the read_csv role of pandas autonomously from loading the CSV file and the parameters which can exist customized to go improve output from the read_csv function.
pandas.read_csv
- Syntax: pandas.read_csv( filepath_or_buffer, sep, header, index_col, usecols, prefix, dtype, converters, skiprows, skiprows, nrows, na_values, parse_dates)Purpose: Read a comma-separated values (csv) file into DataFrame. Also supports optionally iterating or breaking the file into chunks.
- Parameters:
- filepath_or_buffer : str, path object or file-like object Any valid string path is acceptable. The string could be a URL too. Path object refers to os.PathLike. File-like objects with a read() method, such as a filehandle (e.g. via built-in open office) or StringIO.
- sep : str, (Default ',') Separating boundary which distinguishes between whatever two subsequent information items.
- header : int, list of int, (Default 'infer') Row number(due south) to utilise every bit the cavalcade names, and the first of the information. The default behavior is to infer the column names: if no names are passed the behavior is identical to header=0 and cavalcade names are inferred from the first line of the file.
- names : array-like List of column names to utilise. If the file contains a header row, so y'all should explicitly pass header=0 to override the column names. Duplicates in this list are not immune.
- index_col : int, str, sequence of int/str, or False, (Default None) Cavalcade(s) to employ as the row labels of the DataFrame, either given as string name or column index. If a sequence of int/str is given, a MultiIndex is used.
- usecols : list-like or callable Return a subset of the columns. If callable, the callable office will be evaluated against the column names, returning names where the callable role evaluates to True.
- prefix : str Prefix to add to column numbers when no header, e.g. '10' for X0, X1
- dtype : Type name or dict of cavalcade -> type Data type for information or columns. E.g. {'a': np.float64, 'b': np.int32, 'c': 'Int64'} Use str or object together with suitable na_values settings to preserve and not interpret dtype.
- converters : dict Dict of functions for converting values in certain columns. Keys tin can either be integers or column labels.
- skiprows : list-like, int or callable Line numbers to skip (0-indexed) or the number of lines to skip (int) at the start of the file. If callable, the callable function volition be evaluated against the row indices, returning True if the row should be skipped and False otherwise.
- skipfooter : int Number of lines at bottom of the file to skip
- nrows : int Number of rows of file to read. Useful for reading pieces of large files.
- na_values : scalar, str, list-like, or dict Boosted strings to recognize as NA/NaN. If dict passed, specific per-cavalcade NA values. By default the following values are interpreted equally NaN: '', '#Due north/A', '#North/A Northward/A', '#NA', '-1.#IND', '-1.#QNAN', '-NaN', '-nan', '1.#IND', '1.#QNAN', '', 'N/A', 'NA', 'Null', 'NaN', 'north/a', 'nan', 'goose egg'.
- parse_dates : bool or listing of int or names or list of lists or dict, (default Simulated) If set to True, will try to parse the index, else parse the columns passed
- Returns: DataFrame or TextParser, A comma-separated values (CSV) file is returned as a two-dimensional information structure with labeled axes. _For total list of parameters, refer to the offical documentation
Reading CSV file
The pandas read_csv function can be used in dissimilar ways as per necessity like using custom separators, reading just selective columns/rows then on. All cases are covered below ane later on some other.
Default Separator
To read a CSV file, call the pandas function read_csv() and laissez passer the file path as input.
Step 1: Import Pandas
import pandas as pd Step 2: Read the CSV
# Read the csv file df = pd.read_csv("data1.csv") # First 5 rows df.head() 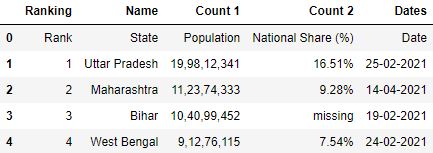
Different, Custom Separators
By default, a CSV is seperated by comma. But you tin can apply other seperators as well. The pandas.read_csvpart is non express to reading the CSV file with default separator (i.e. comma). It can be used for other separators such as ;, | or :. To load CSV files with such separators, the sep parameter is used to pass the separator used in the CSV file.
Permit's load a file with | separator
# Read the csv file sep='|' df = pd.read_csv("data2.csv", sep='|') df 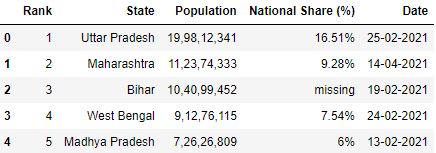
Set any row as column header
Let'south see the data frame created using the read_csv pandas function without whatever header parameter:
# Read the csv file df = pd.read_csv("data1.csv") df.caput() 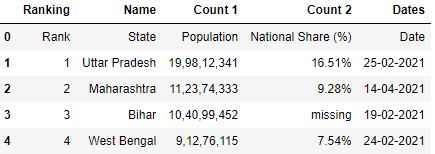
The row 0 seems to be a improve fit for the header. It tin can explain better about the figures in the table. You can brand this 0 row as a header while reading the CSV by using the header parameter. Header parameter takes the value every bit a row number.
Note: Row numbering starts from 0 including column header
# Read the csv file with header parameter df = pd.read_csv("data1.csv", header=1) df.head() 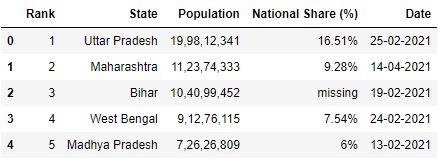
Renaming column headers
While reading the CSV file, you can rename the column headers by using the names parameter. The names parameter takes the list of names of the column header.
# Read the csv file with names parameter df = pd.read_csv( "data.csv" , names=[ 'Ranking' , 'ST Name' , 'Popular' , 'NS' , 'D' ]) df.head() 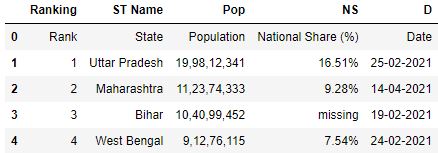
To avoid the old header existence inferred as a row for the data frame, y'all tin can provide the header parameter which will override the quondam header names with new names.
# Read the csv file with header and names parameter df = pd.read_csv( "data.csv" , header=0, names=[ 'Ranking' , 'ST Name' , 'Pop' , 'NS' , 'D' ]) df.head() 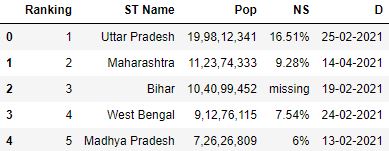
Loading CSV without column headers in pandas
In that location is a chance that the CSV file y'all load doesn't accept any column header. The pandas will make the showtime row every bit a column header in the default case.
# Read the csv file df = pd.read_csv("data3.csv") df.head() 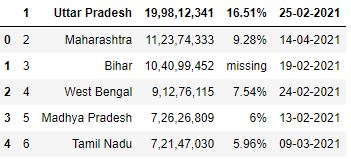
To avoid whatever row being inferred equally column header, you can specify header equally None. It will force pandas to create numbered columns starting from 0.
# Read the csv file with header=None df = pd.read_csv("data3.csv", header=None) df.head() 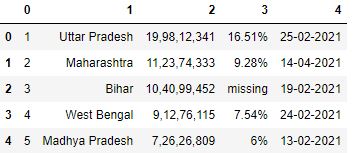
Adding Prefixes to numbered columns
You can also requite prefixes to the numbered cavalcade headers using the prefix parameter of pandas read_csv office.
# Read the csv file with header=None and prefix=column_ df = pd.read_csv("data3.csv", header=None, prefix='column_') df.head() 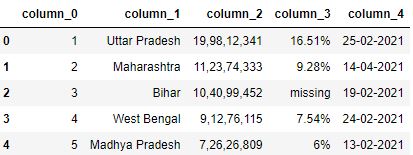
Gear up any column(south) every bit Alphabetize
Past default, Pandas adds an initial alphabetize to the data frame loaded from the CSV file. Yous can control this behavior and make any cavalcade of your CSV as an index by using the index_col parameter.
It takes the name of the desired cavalcade which has to be made as an index.
Case 1: Making 1 cavalcade as index
# Read the csv file with 'Rank' equally index df = pd.read_csv("data.csv", index_col='Rank') df.head() 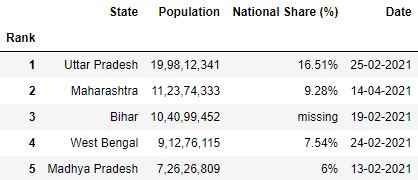
Case ii: Making multiple columns as index
For two or more columns to be made as an index, laissez passer them equally a listing.
# Read the csv file with 'Rank' and 'Date' every bit index df = pd.read_csv("data.csv", index_col=['Rank', 'Date']) df.head() 
Selecting columns while reading CSV
In practice, all the columns of the CSV file are not important. You can select but the necessary columns after loading the file but if yous're enlightened of those beforehand, y'all can save the infinite and time.
usecols parameter takes the list of columns you want to load in your data frame.
Selecting columns using list
# Read the csv file with 'Rank', 'Engagement' and 'Population' columns (list) df = pd.read_csv("information.csv", usecols=['Rank', 'Date', 'Population']) df.caput() 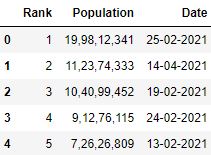
Selecting columns using callable functions
usecols parameter tin can also take callable functions. The callable functions evaluate on column names to select that specific column where the office evaluates to True.
# Read the csv file with columns where length of cavalcade name > 10 df = pd.read_csv("data.csv", usecols=lambda x: len(x)>10) df.caput() 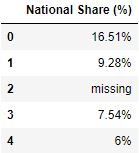
Selecting/skipping rows while reading CSV
You can skip or select a specific number of rows from the dataset using the pandas.read_csv function. At that place are iii parameters that can practise this task: nrows, skiprows and skipfooter.
All of them accept unlike functions. Allow's hash out each of them separately.
A. nrows : This parameter allows you to control how many rows you desire to load from the CSV file. It takes an integer specifying row count.
# Read the csv file with 5 rows df = pd.read_csv("data.csv", nrows=five) df 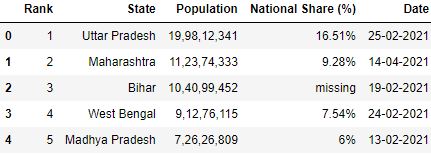
B. skiprows : This parameter allows you to skip rows from the get-go of the file.
Skiprows by specifying row indices
# Read the csv file with first row skipped df = pd.read_csv("data.csv", skiprows=1) df.caput() 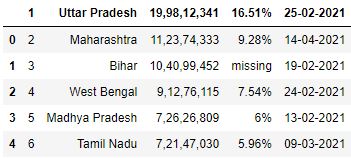
Skiprows by using callback function
skiprows parameter can as well have a callable function equally input which evaluates on row indices. This means the callable role will bank check for every row indices to decide if that row should be skipped or non.
# Read the csv file with odd rows skipped df = pd.read_csv("information.csv", skiprows=lambda x: ten%ii!=0) df.caput() 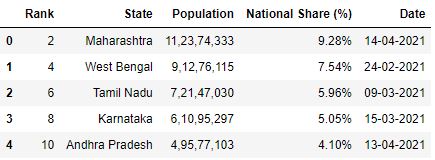
C. skipfooter : This parameter allows you to skip rows from the terminate of the file.
# Read the csv file with 1 row skipped from the end df = pd.read_csv("data.csv", skipfooter=1) df.tail() 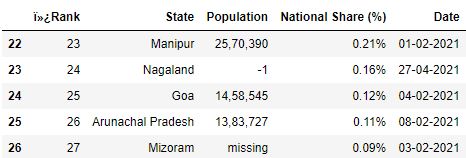
Changing the information type of columns
You can specify the data types of columns while reading the CSV file. dtype parameter takes in the lexicon of columns with their information types defined. To assign the data types, you can import them from the numpy package and mention them confronting suitable columns.
Data Blazon of Rank before change
# Read the csv file df = pd.read_csv("information.csv") # Brandish datatype of Rank df.Rank.dtypes dtype ('int64') Data Type of Rank later modify
# import numpy import numpy as np # Read the csv file with data type specified for Rank. df = pd.read_csv("information.csv", dtype={'Rank':np.int8}) # Display informationtype of rank df.Rank.dtypes dtype ('int8') Parse Dates while reading CSV
Engagement time values are very crucial for data analysis. You can convert a column to a datetime type cavalcade while reading the CSV in two ways:
Method 1. Make the desired cavalcade as an index and pass parse_dates=True
# Read the csv file with 'Date' as index and parse_dates=True df = pd.read_csv("data.csv", index_col='Engagement', parse_dates=True, nrows=5) # Display alphabetize df.index DatetimeIndex(['2021 -02 -25', '2021 -04 -fourteen', '2021 -02 -19', '2021 -02 -24', '2021 -02 -13'], dtype='datetime64[ns]', name='Appointment', freq=None) Method ii. Pass desired cavalcade in parse_dates every bit list
# Read the csv file with parse_dates=['Date'] df = pd.read_csv("data.csv", parse_dates=['Date'], nrows=5) # Display datatypes of columns df.dtypes Rank int64 Country object Population object National Share (%) object Appointment datetime64[ns] dtype: object Adding more NaN values
Pandas library tin handle a lot of missing values. But there are many cases where the data contains missing values in forms that are not present in the pandas NA values list. It doesn't empathise 'missing', 'not establish', or 'not available' as missing values.
So, you lot need to assign them equally missing. To do this, use the na_values parameter that takes a list of such values.
Loading CSV without specifying na_values
# Read the csv file df = pd.read_csv("data.csv", nrows=5) df 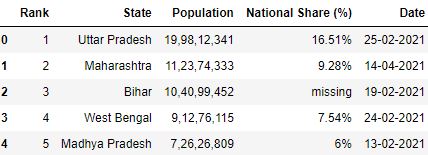
Loading CSV with specifying na_values
# Read the csv file with 'missing' as na_values df = pd.read_csv("information.csv", na_values=['missing'], nrows=5) df 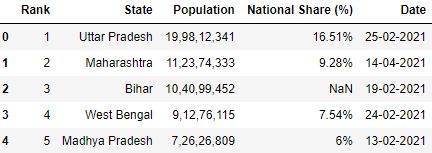
Convert values of the cavalcade while reading CSV
You tin can transform, modify, or catechumen the values of the columns of the CSV file while loading the CSV itself. This tin be washed by using the converters parameter. converters takes in a lexicon with keys equally the column names and values are the functions to exist practical to them.
Let'due south convert the comma seperated values (i.eastward 19,98,12,341) of the Population column in the dataset to integer value (199812341) while reading the CSV.
# Role which converts comma seperated value to integer toInt = lambda x: int(x.replace(',', '')) if x!='missing' else -1 # Read the csv file df = pd.read_csv("data.csv", converters={'Population': toInt}) df.head() 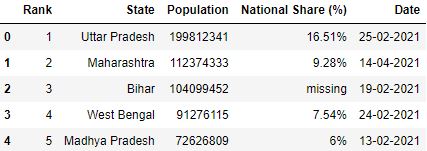
Applied Tips
- Before loading the CSV file into a pandas data frame, e'er have a skimmed await at the file. It volition assistance you lot estimate which columns you should import and make up one's mind what data types your columns should have.
- You should also lookout for the total row count of the dataset. A system with iv GB RAM may not be able to load 7-8M rows.
Test your knowledge
Q1: You cannot load files with the $ separator using the pandas read_csv part. Truthful or Simulated?
Answer:
Reply: False. Because, you can use sep parameter in read_csv role.
Q2: What is the use of the converters parameter in the read_csv part?
Answer:
Respond: converters parameter is used to modify the values of the columns while loading the CSV.
Q3: How will you make pandas recognize that a particular column is datetime type?
Reply:
Answer: Past using parse_dates parameter.
Q4: A dataset contains missing values no, not bachelor, and '-100'. How will you lot specify them every bit missing values for Pandas to correctly interpret them? (Assume CSV file name: example1.csv)
Answer:
Answer: By using na_values parameter.
import pandas as pd df = pd.read_csv("example1.csv", na_values=['no', 'not available', '-100']) Q5: How would you lot read a CSV file where,
- The heading of the columns is in the 3rd row (numbered from 1).
- The terminal 5 lines of the file have garbage text and should be avoided.
- Only the column names whose first letter starts with vowels should exist included. Assume they are one give-and-take only.
(CSV file name: example2.csv)
Respond:
Answer:
import pandas as pd colnameWithVowels = lambda x: 10.lower()[0] in ['a', 'e', 'i', 'o', 'u'] df = pd.read_csv("example2.csv", usecols=colnameWithVowels, header=2, skipfooter=5) The article was contributed past Kaustubh K and Shrivarsheni
zimmermanhunce1987.blogspot.com
Source: https://www.machinelearningplus.com/pandas/pandas-read_csv-completed/
0 Response to "How to Read Csv With No Header"
Post a Comment